Help
- I. Formation Mechanism of tsRNA
- II. Confidence Classification of tsRNA-Disease Associations
- III. The usage of tsRNADisease v1.0 database
- IV. Contact Us
- V. Note
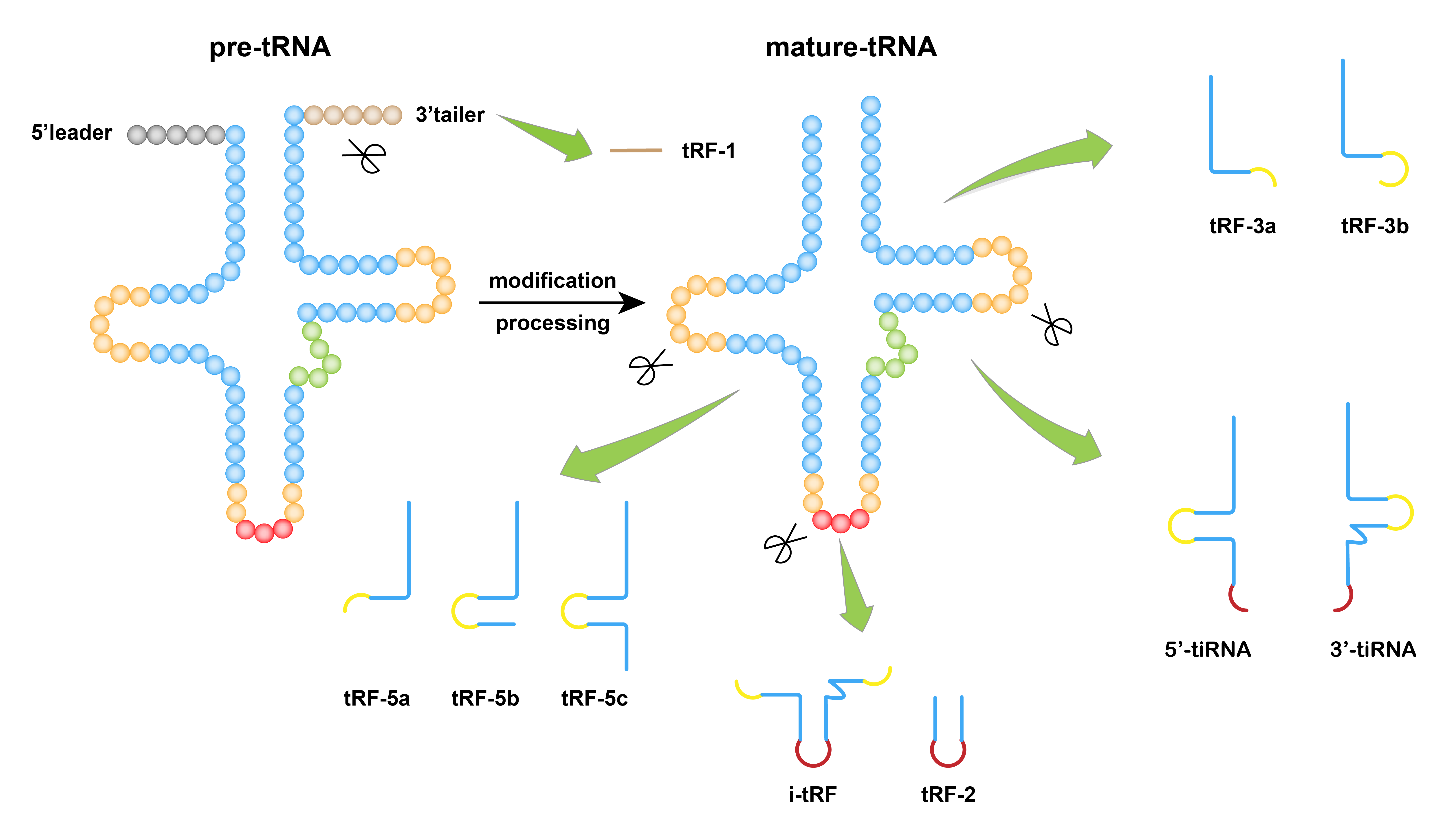
I. Formation Mechanism of tsRNA
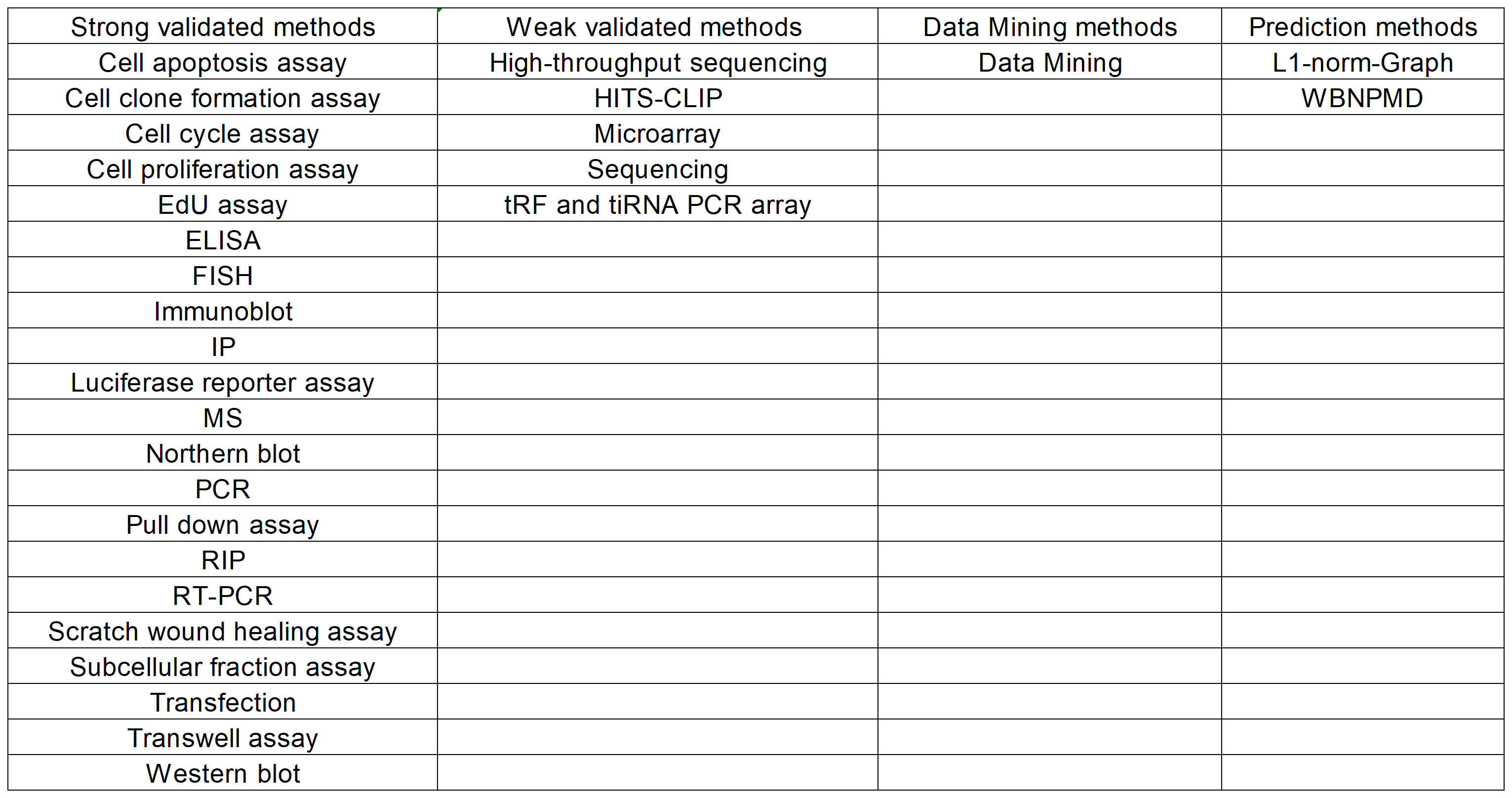
II. Confidence Classification of tsRNA-Disease Associations
Based on the supporting methods for each tsRNA-disease associations, we have classified them into four confidence levels:
·Very High: Supported by strong validated methods with functional experiments.
·High: Supported by strong validated methods without functional experiments.
·Median: Supported by weak validated methods other than data mining.
·Low: Supported only by data mining methods.
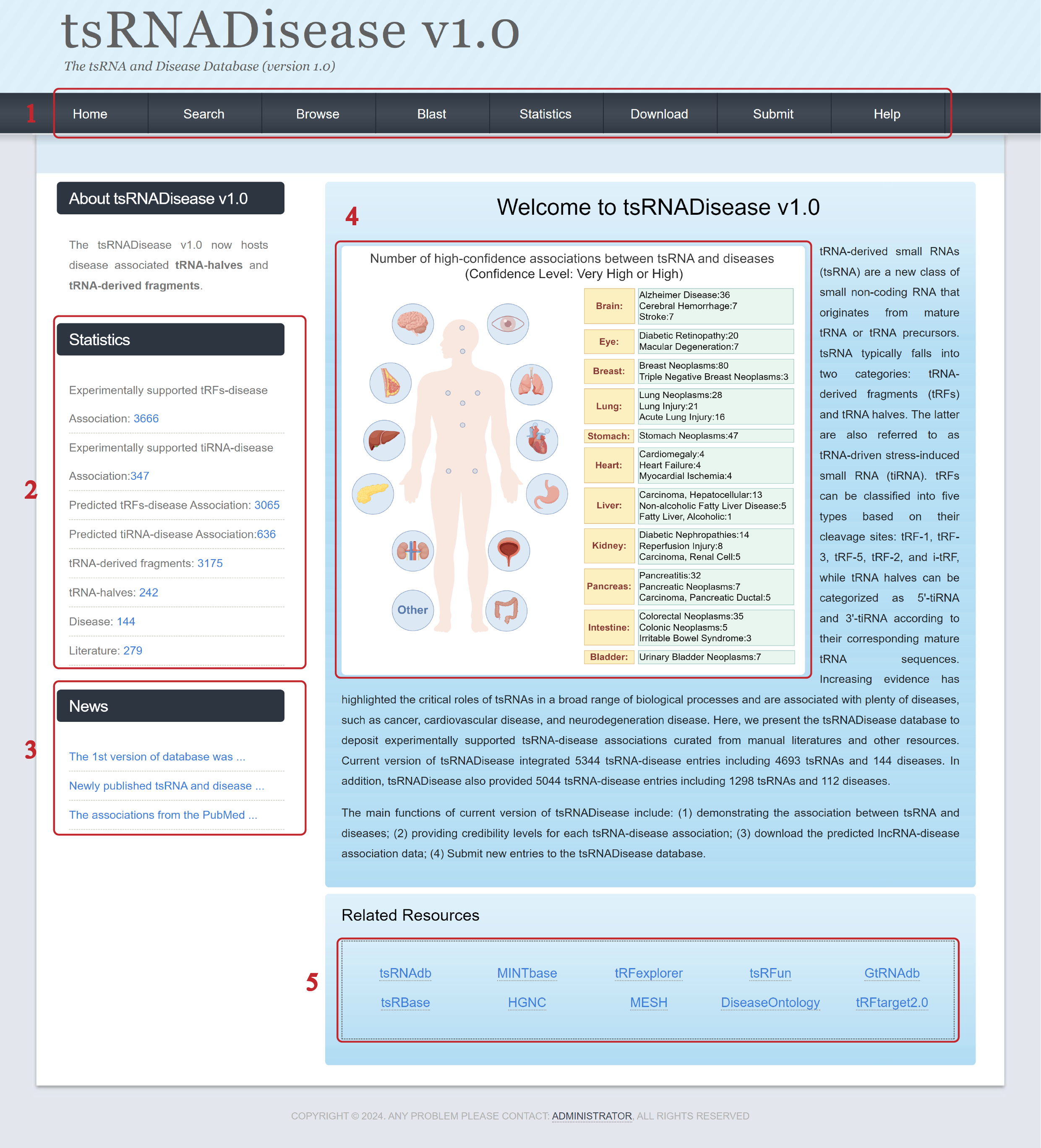
III. The usage of tsRNADisease v1.0 database
1. Overview of tsRNADisease v1.0
1. Main functions of the database are provided in the menu bar.
2. Main statistics of the database.
3. Updated news about the database.
4. Partial tsRNA-disease associations are displayed; more detailed information will be shown when hovering over the organ diagram.
5. Related resources to the database.
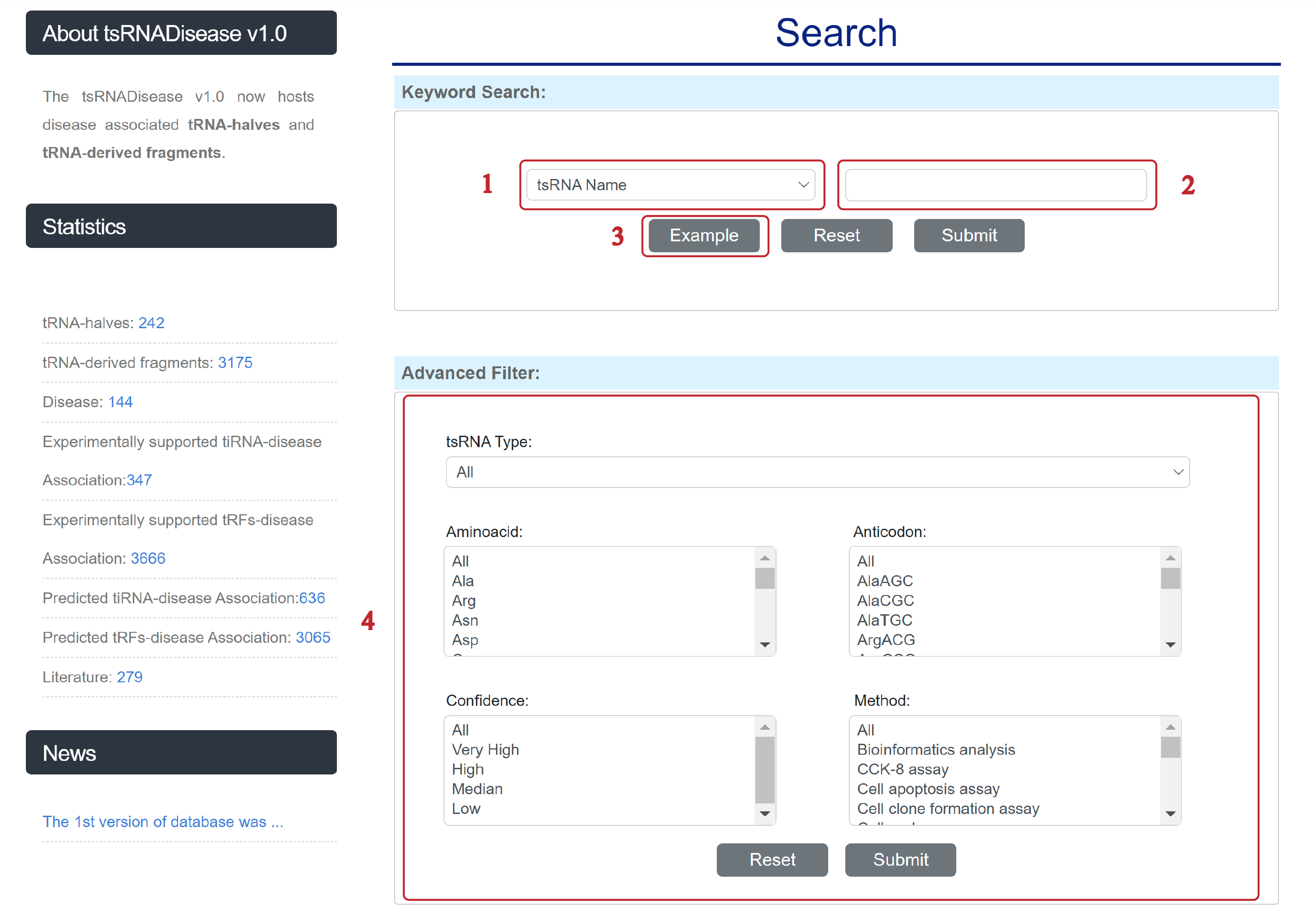
2. Search Database
The Search page is displayed as follows.
1. Select a category: Four choices are provided.
2. Input a keyword corresponding to the selected category.
3. Click to view example.
4. Five categories provided to filter results.
① Use Disease Ontology/MeSH/NCBI Gene to normalize your input information.
② If you got no result or too few results, you may shorten your keywords.
3. Search Result
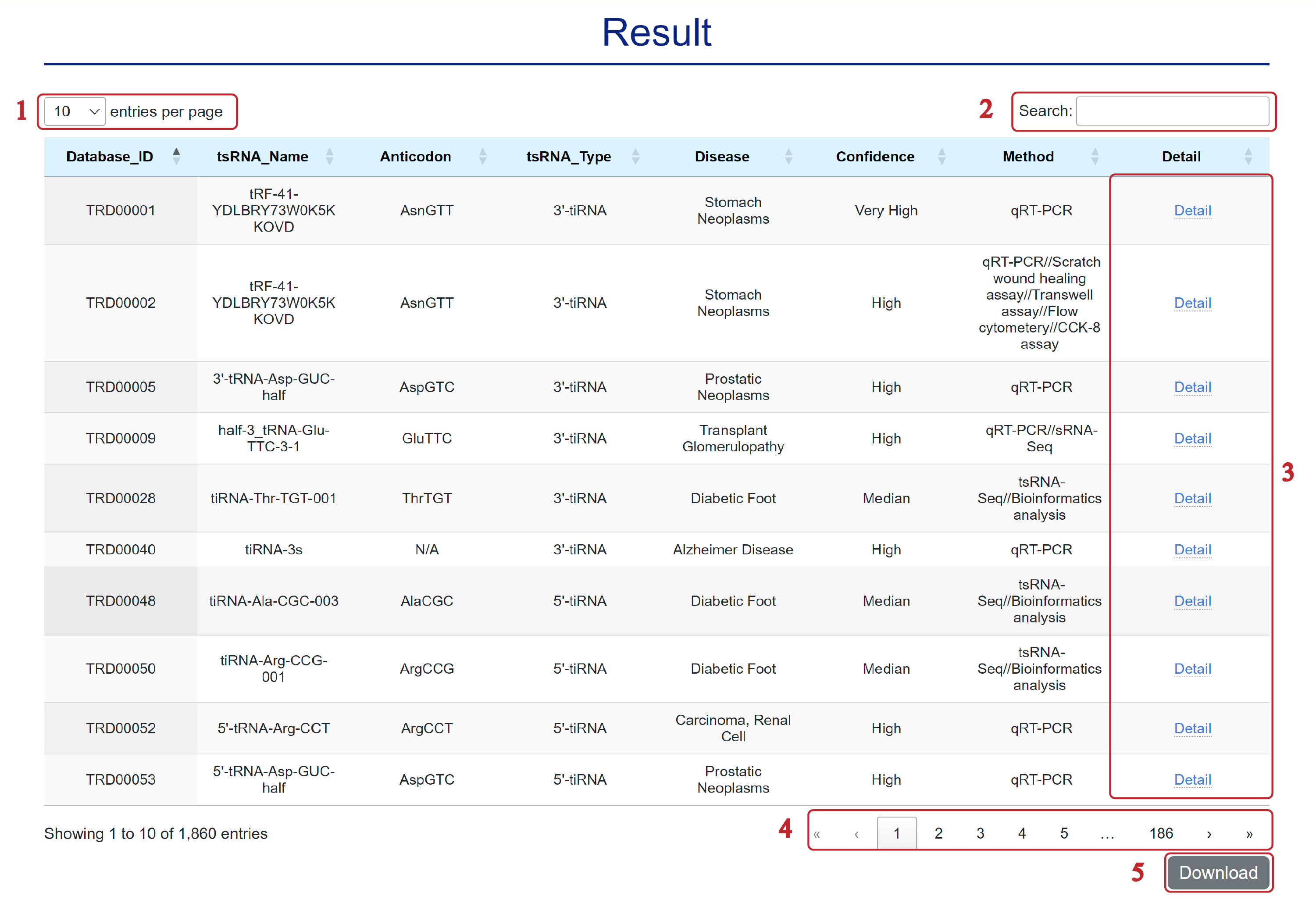
The Result page is displayed as follows.
In the Result page, all associations are listed with basic information including tsRNA_Name, Anticodon, tsRNA_Type, Disease, Confidence, Method and Detail.
1. Select the number of entries per page.
2. Search the table.
3. Click to link to the Entry Detail page.
4. Click to turn the page.
5. Click to download the table.
4. Browse Database
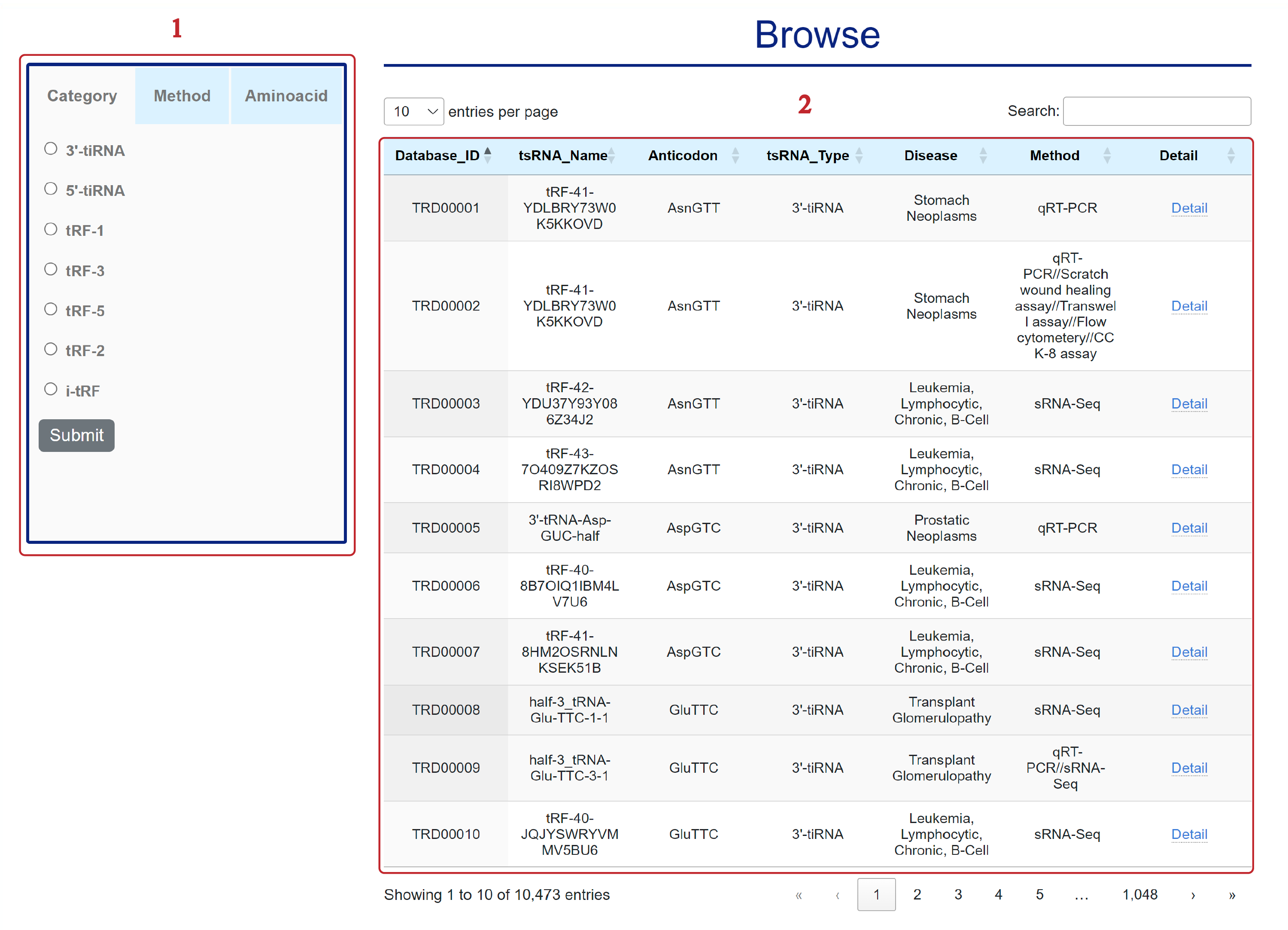
The Browse page is displayed as follows.
Associations can be displayed selectively according to selected conditions.
1. Browse by different categories.
2. Database items.
5. Blast
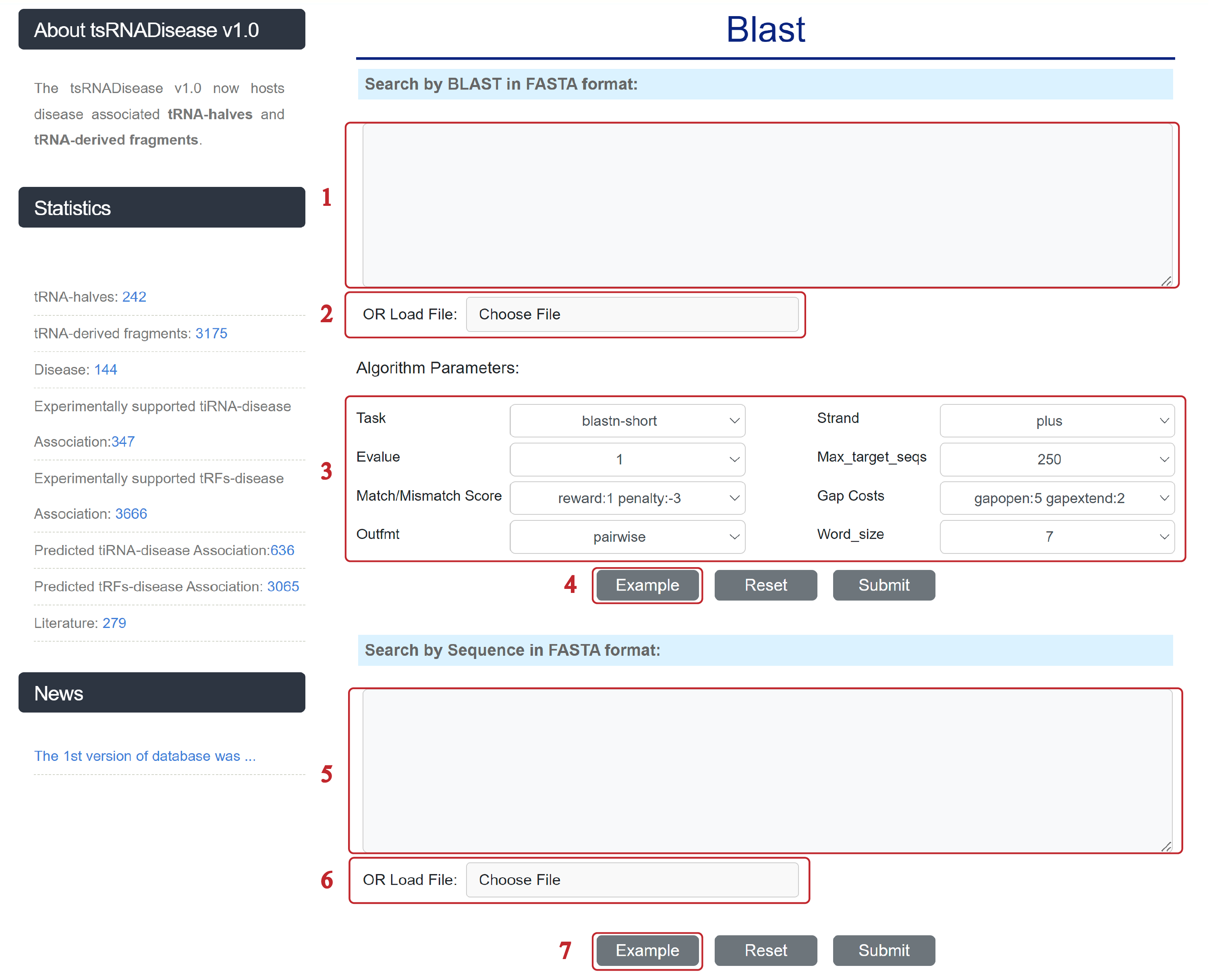
The Blast page is displayed as follows.
1&5. Input tsRNA sequences.
2&6. Upload a text file containing tsRNA sequences.
3. Select the appropriate parameters.
4&7. Click to view example.
6. Entry Detail
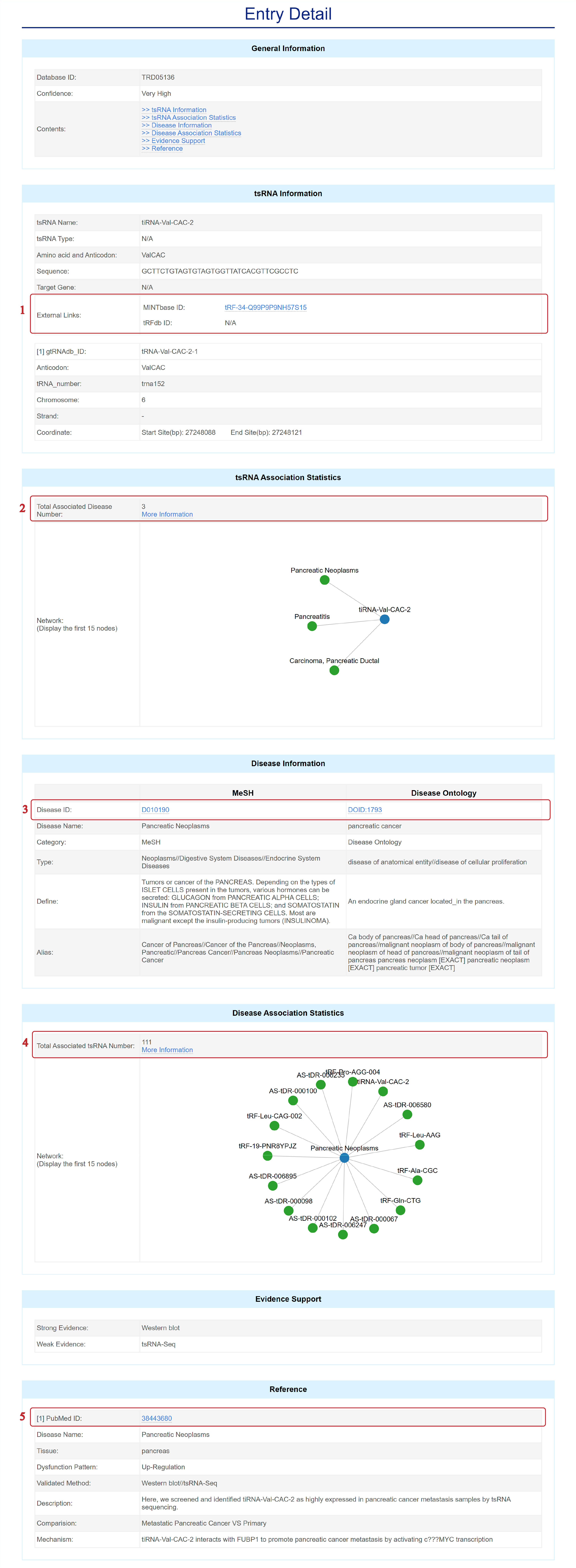
The Entry Detail page is displayed as follows.
In the Entry Detail page, you can get information including General Information, ncRNA Information, Regulatory Relationship, Disease Information, Evidence Support and Reference.
1. Click MINTbase ID, tRFdb ID to get more detailed information in external databases.
2. Click to view more associated diseases.
3. Browse Disease Ontology ID/MeSH to get more detailed information in Disease Ontology/MeSH.
4. Click to view more associated tsRNAs.
5. Click PubMed ID to get more detailed information in NCBI.
IV. Contact Us
This database is hosted by the following organizations:
1.Computational Genetics Group, Institute of Biomedical Sciences, Fudan University.
2.Anhui Province Key Laboratory of Non-coding RNA Basic and Clinical Transformation, Tissue Bank, The First Affiliated Hospital of Wannan Medical College (Yijishan Hospital of Wannan Medical College), Central Laboratory, The First Affiliated Hospital of Wannan Medical College (Yijishan Hospital of Wannan Medical College), Wuhu, Anhui, China.
If you have any questions or suggestions about our database, please don't hesitate to contact us.
Database Administrator: zhenyang@fudan.edu.cn
Member:Hui Yang yanghui0203@wnmc.edu.cn
Member:zhusy22@m.fudan.edu.cn
V. Note
The diagram titled 'Number of high-confidence associations between tsRNA and diseases (Confidence Level: Very High or High)' on the Home page was created by Figdraw (https://www.figdraw.com).